CellNet is a network biology-based computational platform that more accurately assesses the fidelity of cellular engineering than existing methodologies and generates hypotheses for improving cell derivations. CellNet is based on the reconstruction of cell type specific gene regulatory networks (GRNs), which we performed using publicly available microarray data on 21 cell and tissue types from both human and mouse samples. Code to run CellNet locally is available for download on this site. You can also analyze your data through our our servers, where you can also explore predicted transcriptional targets of over 1,200 mouse or human transcriptional regulators.
CellNet was written by Patrick Cahan with input from Hu Li, Samantha Morris, and Edroaldo Lummertz da Rocha.
It is provided under the OSI-approved Artistic License (version 2.0).
Release 0.1 -- 08-14-2014
The first version of CellNet is now available. This software a beta release, meaning that it is a work in progress, and new features will be added in the near future. To report a bugs or to discuss this software (but not the web application, please), go to the CellNet user group.
Requirements
CellNet was written in R. The following R packages, available through CRAN, are required:
- ggplot2
- gplots
- randomForest
- igraph
You can install them packages from within an R session by typing:
> install.packages(c("ggplot2", "gplots", "randomForest", "igraph"))
You will also need to install the affy package from Bioconductor. You can install it from within an R session by typing:
> source("http://bioconductor.org/biocLite.R")
> biocLite("affy")
You will also need to install the following Bioconductor annotation packages. Please note that you must download these particular versions in order for CellNet to run properly:
- org.Mm.eg.db_2.8.0
- org.Hs.eg.db_2.8.0
- mouse4302.db
- mouse4302cdf_2.11.0
- hgu133plus2.db
- hgu133plus2cdf
- hugene10sttranscriptcluster.db_8.0.1
- hugene10stv1cdf
- mogene10sttranscriptcluster.db_8.0.1
- mogene10stv1cdf
We recommend that you install these packages to directory where they will not be overwritten. For example, we install these in the directory ~/sample_packages/install_dir. After you have downloaded these files, you can install them by typing at the shell:
R CMD INSTALL -l ~/sample_packages/install_dir package name
Installation
To install CellNet, download the compressed package, then at the shell type:
R CMD INSTALL -l ~/sample_packages/install_dir cellnetr.0.1.tar.gz
Finally, you need to download CellNet Objects, one per platform, which contain the training data, classifiers, and gene regulatory networks
The directory where you place these files will be referred to as 'path_CN_obj' in the code snippets below.Setting up R environment for CellNet
# Set your library path so that it points to the correct platform and annotation libraries:
.libPaths("~/sample_packages/install_dir")
# load cellnet
library("cellnetr")
# load appropriate gene annotation sources
# mouse
library("org.Mm.eg.db")
# if human
# library("org.Hs.eg.db")
# load platform annotation
# mouse4302
library("mouse4302cdf");
library("mouse4302.db");
# mogene10
# library("mogene10sttranscriptcluster.db");
# library("mogene10stv1cdf");
# hgu133plus2
# library("hgu133plus2.db");
# library("hgu133plus2cdf");
# hugene10
# library("hugene10sttranscriptcluster.db");
# library("hugene10stv1cdf");
# set up path the CellNet objects containing the classifiers, GRNs, etc
# Edit this so that it points to the path where you saved the CellNet objects
path_CN_obj<-"~/CellNet/CN_Objects/";
# change to reflect your platform
myPlatform<-"mouse4302"
# name of the column in the sample data table that indicates experimental groups or replicates
# set to "", or "sample_name" if there are no replicates
cName<-"description1"
# target tissue or cell type.
targetCT<-"hspc"
Load data and run CellNet
# Load sample table and fix the data file names
# replace 'sampleTab.csv' with your sample table name. See [Step 2 here](http://dev.cellnet.hms.harvard.edu/run/) for a description of the sample table format and an example table.
stQuery<-expr_readSampTab("sampleTab.csv");
stQuery<-geo_fixNames(stQuery);
# load and normalize expression data
expQuery<-Norm_cleanPropRaw(stQuery, "mouse4302");
# load right CellNet object
cnObjName<-switch(myPlatform,
mouse4302 = paste(path_CN_obj, "cnProc_mouse4302_062414.R",sep=''),
mogene10stv1 = paste(path_CN_obj, "cnProc_mogene_062414.R",sep=''),
hgu133plus2 = paste(path_CN_obj, "cnProc_Hg1332_062414.R",sep=''),
hugene10stv1 = paste(path_CN_obj, "cnProc_Hugene_062414.R",sep=''));
cnProc<-utils_loadObject(cnObjName);
# Run CellNet
tmpAns<-cn_apply(expQuery, stQuery, cnProc, dLevelQuery=cName);
# Score transcription factors
tfScores<-cn_nis_all(tmpAns, cnProc, targetCT);
Visualize the results
# Classification heatmap
cn_hmClass(tmpAns);
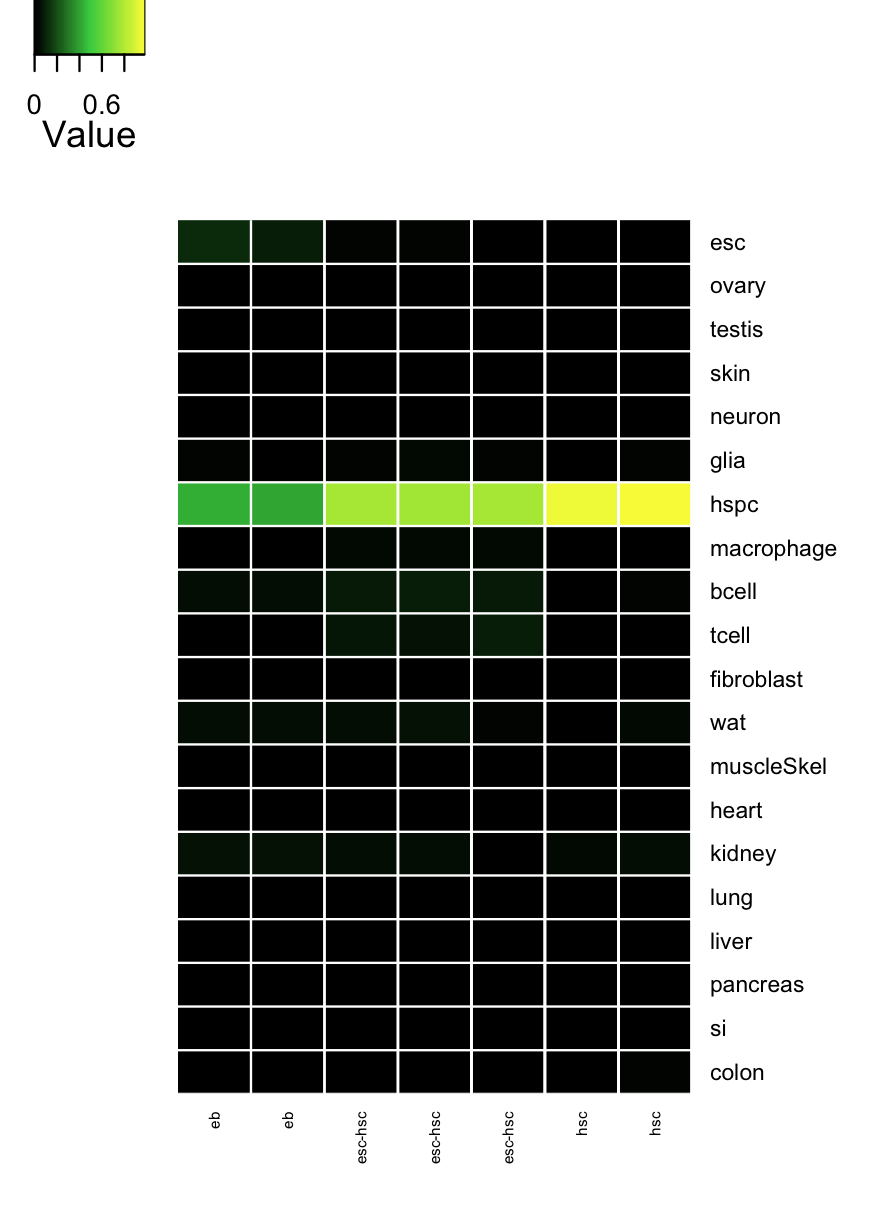
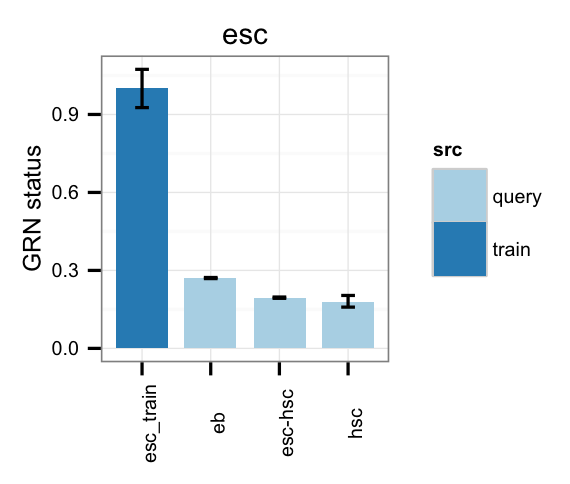
# Gene regulatory network status of starting cell type (esc) GRN
cn_barplot_grnSing(tmpAns, cnProc, "esc", c("esc"), bOrder=NULL, norm=T);
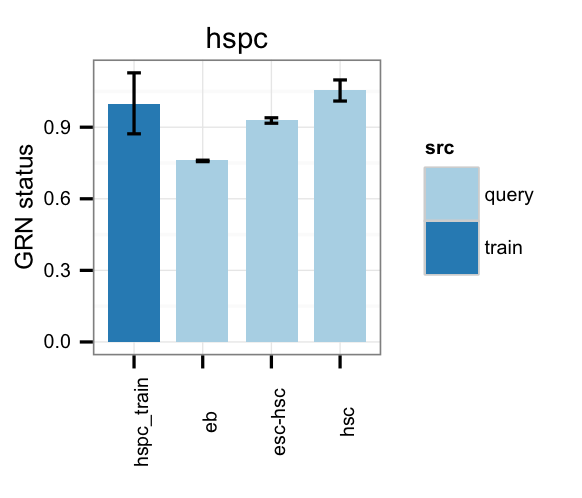
# Gene regulatory network status of target cell type (hspc) GRN
cn_barplot_grnSing(tmpAns, cnProc, "hspc", c("hspc"), bOrder=NULL, norm=T);
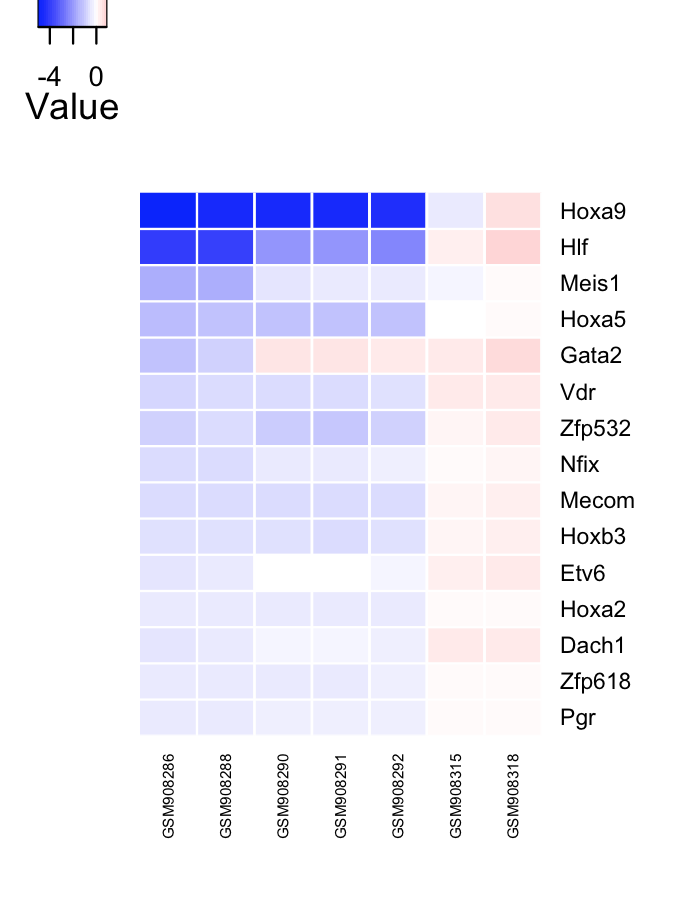
# Network influence score of HSPC GRN transcriptional regulators.
# factor heatmap
cn_plotnis(tfScores, limit=15);
# Gene expression in the training data set
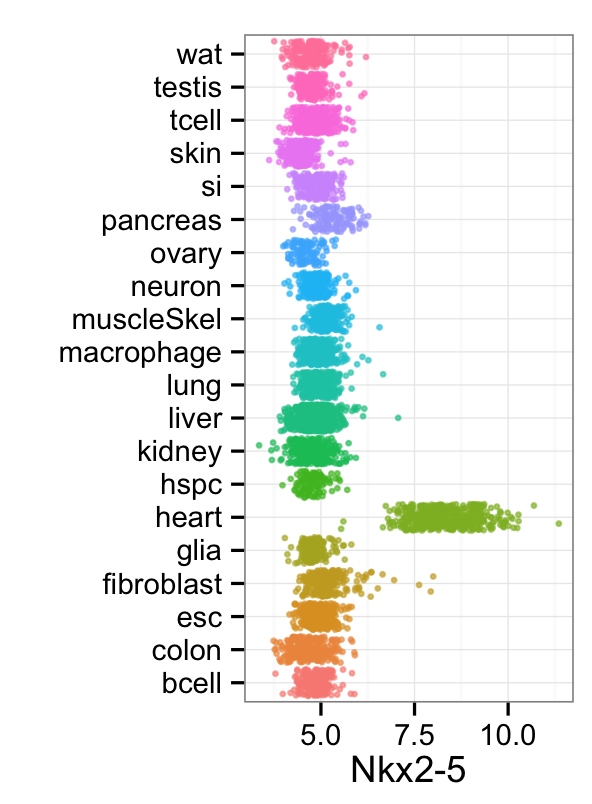
mp_rainbowPlot(cnProc[['expTrain']],cnProc[['stTrain']],"Meis1", dLevel="description1")
Features to be added in the near future
- Adding a cell or tissue type
- Adding a new platform
- Adding a new species
- Reconstructing GRNs de novo
- Identifying cell and tissue specific sub-networks